RNA Sequencing and Basic MATLAB Data Preprocessing Guide
Written on
Chapter 1: Introduction to RNA Sequencing
In my research lab focused on fundamental biological processes, we utilize yeast as a model organism for various experiments. Recently, a physicist friend and technology marketing executive joined me in this endeavor. These notes serve as an informal introduction to RNA sequencing, aimed at helping him and others interested in transitioning to a biotechnology lab.
1. Understanding RNA Sequencing for Protein Regulation…
My focus lies in examining the regulation of specific genes. One effective method to explore gene regulation is through RNA sequencing, which allows us to analyze the RNA produced by a gene.
Earlier, we discussed how genes (DNA) encode the information required for protein synthesis. All living organisms, from bacteria to humans, utilize messenger RNA (mRNA) as an intermediary that carries this information from DNA to proteins. Notably, mRNA has gained prominence as the foundation for the first successful COVID-19 vaccines.
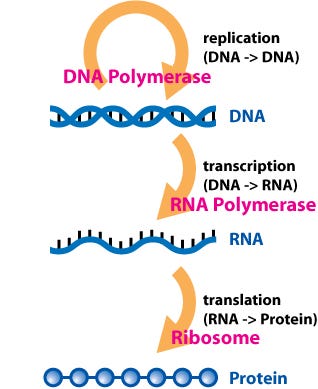
mRNA provides additional insights into protein production, including its synthesis location, timing, and quantity. It also carries regulatory instructions for protein synthesis, while some directives remain encoded in DNA. The advancements in sequencing technology have made it increasingly accessible to sequence RNA, providing crucial data on how cells manage protein synthesis.
2. A Closer Look at RNA…
RNA is a molecule structurally akin to DNA. While DNA comprises four types of nucleotides known as dA, dT, dG, and dC, RNA is formed from A, U, G, and C. The "d" prefix signifies deoxyribonucleic acid, distinguishing it from ribonucleic acid.
An enzyme called RNA polymerase reads DNA and synthesizes RNA by following complementary base-pairing rules. For instance, if RNA polymerase encounters a dA, it will produce a U, as follows:
3. RNA Isolation for Sequencing…
The initial step in RNA sequencing involves isolating RNA from cells, a process we previously explored in my article on COVID-19 nasal swab tests.
Once isolated, we convert RNA to DNA for two key reasons: DNA's stability simplifies handling and sequencing technologies predominantly operate on DNA. During this conversion, specific sequences, such as a barcode for identification, are attached to the RNA.
The enzyme reverse transcriptase (RT) facilitates this conversion, reading RNA and synthesizing complementary DNA (cDNA) using the same base-pairing principles.
4. RNA Sequencing Techniques…
Modern sequencing technologies, like those from Illumina, employ DNA polymerase to read cDNA and synthesize complementary DNA.
This process involves labeling each nucleotide with a unique fluorophore, allowing machines to record sequences as they are synthesized. This parallel processing occurs across millions of RNA molecules simultaneously, employing techniques such as paired-end sequencing to optimize data collection.
5. Analyzing Sequencing Data…
The output from sequencers consists of text files containing extensive data, typically organized into three components: a unique identifier, the RNA nucleotide sequence, and a quality score.
For example, a single read might look like this:
@GWNJ-1013:136:GW2102251027th.Miseq:1:2101:30219:1000 2:N:0:TCTCGCGC+TTCCGCTT
CAGGAGCGATGAGAAGCGCCTTTCGACCACTTTGGTAAAAATCTAGTTTTTCCAAATCAGTGGAGACAAGAAGACGATAATCAAAGACCGAAACGTCGAAAACAAACACTAGCAGCTAAATCTAGTGTTGTTCTTGCTTTGCAAACACCGTTCTATGGCTCAAAAGCGAGGAGTCAGAGACAATCTACAGAACAGTTTAAACGCCAAAAAAAAAACGGGCAATAAGCAGTAGCTATCGAGCAGAAGTAAT
FFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFF...
6. MATLAB for Data Processing…
Disclaimer: I am not a programmer. The following sections describe my process rather than serve as a definitive guide.
My first foray into coding was in MATLAB, where I used the fastqread function to import paired-end read files from the sequencing company.
For example, to read the files, I executed:
FASTQStruct1 = fastqread('97–21-K_R1_001.fastq');
FASTQStruct2 = fastqread('97–21-K_R2_001.fastq');
Next, I reversed the complement of one read to ensure proper alignment before merging them.
for n = 1:N2
FASTQStruct2RC(n).Sequence = seqrcomplement(FASTQStruct2(n).Sequence);
end
7. Final Steps and Conclusion…
The final stages of my analysis involved trimming and filtering the sequences based on specific RNA adapters and primers for different yeast species. This resulted in clean, usable data for further experimentation.
In summary, we prepared RNA from various yeast species and sequenced it to analyze a specific gene, utilizing MATLAB for data preprocessing. The key steps included:
- Importing fastq data
- Reverse complementing reads
- Merging sequences
- Filtering based on specific criteria
- Quality checks throughout the process
Thank you for reading. Further articles in this series will cover various topics in molecular biology, biochemistry, and genetics.
